New Pubmed’s interface

PubMed has presented its new interface, cleaner and simpler, optimized for mobile devices and with new search algorithms.
There is one thing that happens on a recurring basis and that is a real slap in the face for me. It turns out that I like to go shopping for food once a week, so I usually go to the hypermarket every Friday. I am a creature of habit that always eats the same things and almost the same days, so I go swift and fast through the aisles of the hyper throwing things in the shopping cart so I have the matter settled in the twinkling of an eye.
The problem is that in hypermarkets they have the bad habit of periodically changing foods sites, so you go crazy until you learn its new location again. To cap it all, the first few days foods have been changed, but not yet its information panels, so I have to go around a thousand turns until I find the cans of squid in their ink that, as we all know, are one of our main staple foods.
You will wonder why I tell you all this stuff. As it turns out, the National Library of Medicine (NML) has done a similar thing: now that I had finally managed to learn how the its search engine worked, they go and change it completely.
Of course, it must be said in honor of the truth that NML’s people have not limited themselves to changing the aspects of windows and boxes, but have implemented a radical change with an interface that they define as cleaner and simpler, as well as better adapted to mobile devices, which are increasingly used to do bibliographic searches. But that doesn’t end there: there are a lot of improvements in the algorithms to find the more than 30 million citations that Pubmed includes and, in addition, the platform is hosted in the cloud, promising to be more stable and efficient.
New Pubmed’s interface
The NLM announced the new Pubmed in October 2019 and it will be the default option at the beginning of the year 2020 so, although the legacy version will be available a few more months, we have no choice but to learn how to handle the new version. Let’s take a look.
Although all the functionalities that we know of the legacy version are also present in the new one, the aspect is radically different from the home page, which I show you in the first figure.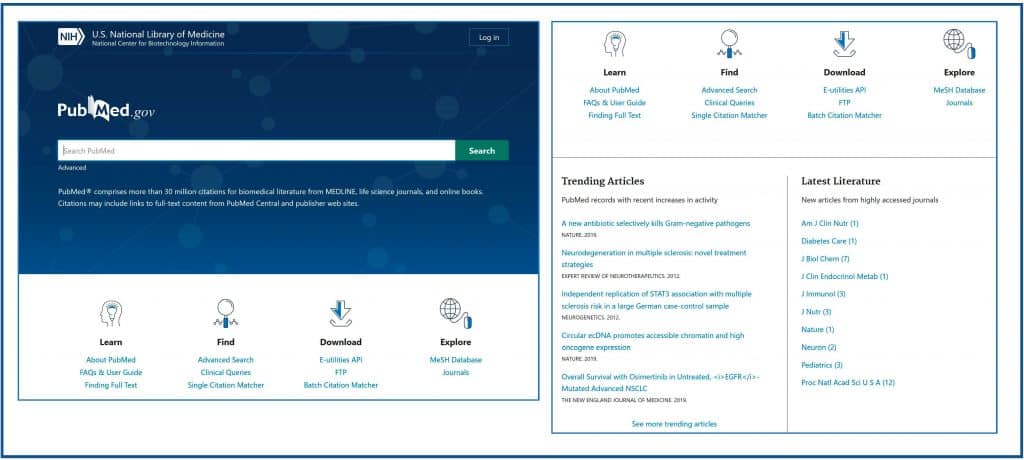
Below we have four sections, including the one that contains help to learn how to use the new version, and that include tools that we already knew, such as “Clinical Queries”, “Single Citation Matcher” or “MeSH Database”. At the time of writing this post, these links direct you to the old versions of the tools, but this will change when the new interface is accessed by default.
Finally, a new component called “Trending Articles” has been added at the bottom. Here are articles of interest, which do not have to be the most recent ones, but those that have aroused interest lately and have been viralized in one way or another. Next to this we have the “Latest Literature” section, where recent articles from high impact journals are shown.
Now let’s see a little how searches are done using the new Pubmed. One of the keys to this update is the simple search box, which has become much smarter by incorporating a series of new sensors that, according to the NLM, try to detect exactly what we want to look for from the text we have inserted.
For example, if we enter information about the author, the abbreviation of the journal and the year of publication, the citation sensor will detect that we have entered basic citation information and will try to find the article we are looking for.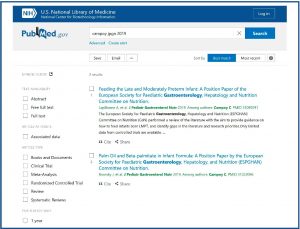
We can also do the search in a more traditional way.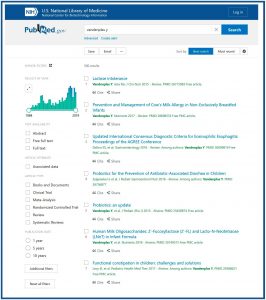
Of course, we can also search by subject. If I type “parkinson” in the search box, Pubmed will make a series of suggestions on similar search terms. If I press “Search”, I get the results of the fourth figure which, as you can see , includes all the results with the related terms.
Let us now turn to the results page, which is also full of surprises. You can see a detail in the fifth figure. Under the search box we have two links: “Advanced”, to access the advanced search, and “Create alert”, so that Pubmed notifies us every time a new related article is incorporated (you already know that for this to be possible we have to create an account in NCBI and enter by pressing the “Login” button at the top; this account is free and saves all our activity in Pubmed for later use).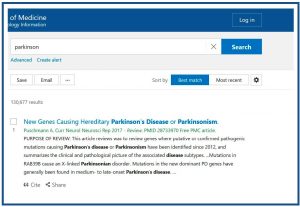
Below these links there are three buttons which allow you to save the search ( “Save”), send it by e-mail (“Email”) and, clicking the three points button, send it to the clipboard or to our bibliography or collections, if we have an NCBI account.
On the right we have the buttons to sort the results. The “Best Match” is one of the new priorities of the NLM, which tries to show us in the first positions the most relevant articles. Anyway, we can sort them in chronological order (“Most recent”), as well as change the way of presenting them by clicking on the gearwheel on the right (in “Summary” or “Abstract” format).
We are going to focus now into the left of the results page. The first thing we see is a graph with the results indexed by year. This graph can be enlarged, which allows us to see the evolution of the number of papers on the subject indexed over time. In addition, we can modify the time interval and restrict the search to what is published in a given period. In the sixth figure I show you how to limit the search to the results of the last 10 years.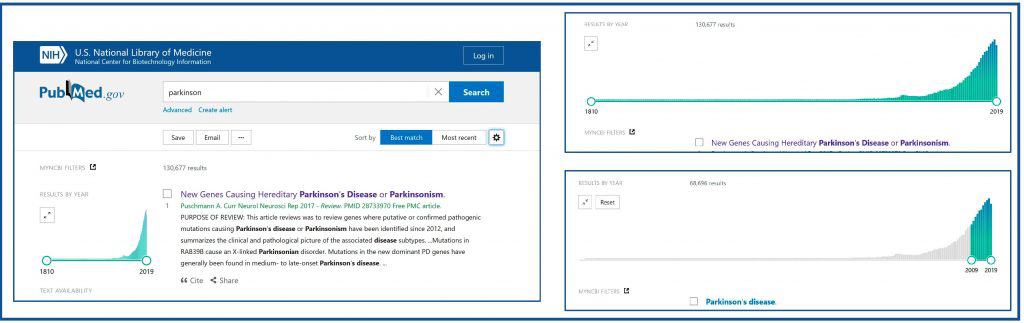
Finally, to the left of the results screen we have the list of filters that we can apply. These can be added or removed in a similar way to how it was done with the legacy version of Pubmed and its operation is very intuitive, so we will not spend more time on them.
If we click on one of the items in the list of results we will access the screen with the text of the paper (seventh figure).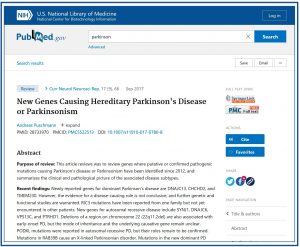
Advanced search
To finish this post, let’s take a look at the new advanced search, which can be accessed by clicking on the “Advanced” link, which will take us to the screen you see in the eighth figure.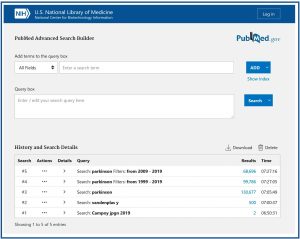
Its operation is very similar to the legacy version. We can add terms with Boolean operators, combine searches, etc. I encourage you to play with the advanced search, the possibilities are endless. The newest part of this tool is the section with the history and the search details (“History and Search Details”) at the bottom. This allows you to keep previous searches and return to them, taking into account that all this is lost when you leave Pubmed, unless you have an NCBI account.
We’re leaving…
And here we end for today. You have seen that these people of the NLM have outdone themselves, putting at our disposal a new tool easier to use, but at the same time, much more powerful and intelligent. Google must be shaking with fear, but don’t worry, it is sure it will invent something to try to prevail.
You can go forgetting about the legacy version, do not wait for it to disappear to start enjoying the new one. We will have to talk about these issues again when new versions of the rest of the tools are established, such as Clinical Queries, but that is another story …